Reaction Workbench’s Mechanism Merge utility provides a powerful and accurate method for combining mechanisms from the same or disparate sources. It provides great flexibility to expert users while providing reasonable default operations for nonexperts. It enables comparison of species and reactions, as well as direct comparison of data provided by different sources for species thermodynamic and transport properties, plus reaction-rate data. In this way, the Mechanism Merge Utility also provides mechanism-comparison capabilities.
The Mechanism Merge utility provides the ability to compare and/or merge two different mechanisms. The comparison step allows detailed analysis of any overlap and differences between the mechanism data, to enable informed decisions to be made during the merge process. This allows users to merge together any two valid mechanisms that are in Ansys Chemkin format. Two options are available for matching the species from the different mechanisms. The choice of which option to use is contingent upon the availability of consistent, canonical SMILES [1] assignments for each species in the two mechanisms being merged:
If every species in the mechanisms to be merged has SMILES™ assigned, the species will be matched between mechanisms using their SMILES assignments.
If any species in any of the mechanisms is missing a SMILES assignment, species will be identified using the species chemical formula (Ansys Chemkin symbolic species name). For example, even though OH and HO are both representing the hydroxyl radical, they would be treated as different species, because their species symbolic name is different. This option is therefore very risky for mechanisms that have not originated from the same source with the same species naming convention.
We strongly recommend option 1, especially if mechanisms are being merged from different sources where naming conventions are likely to differ. For the two sample surrogate mechanisms provided with Reaction Workbench, SMILES strings are already tagged in each of the mechanism files, such that option 1 will always be used. For other mechanisms, you can choose to either employ the utility at your own risk with option 2 or generate the needed SMILES strings for each mechanism to allow use of option 1.
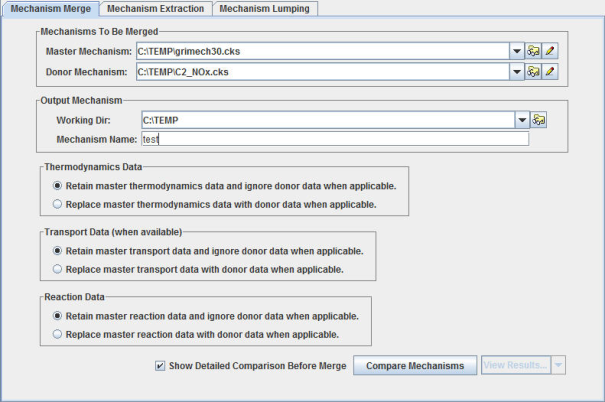
The first step in a mechanism merge is to select or set up chemistry sets for the
mechanisms to be merged. A chemistry set can be created using the pencil icon  next to the mechanism selection pull-down menu. Or you can
browse to an existing chemistry-set file (*.cks file) using the folder search icon
next to the mechanism selection pull-down menu. Or you can
browse to an existing chemistry-set file (*.cks file) using the folder search icon  .
.
To start a merging operation, you need to identify a chemistry set for a full mechanism and for a donor mechanism. The full mechanism is defined as the mechanism that establishes defaults whenever conflicts arise during mechanism comparison, while the donor mechanism will be used as the secondary source of species and reactions that are not found in the full mechanism. So in general, you would identify the mechanism that you trust the most as the "full". During the merge operation, however, you can override these default rules for resolving conflicts by making explicit choices for each conflict, whether it be in species thermodynamic data, species transport data, or reaction rate data. In addition, such options can be specified globally based on the types of data, using the radio buttons on the top-level Mechanism Merge tab. All these options are available on the Mechanism Merge tab, as shown in Figure 2.1: Mechanism Merge in Reaction Workbench .
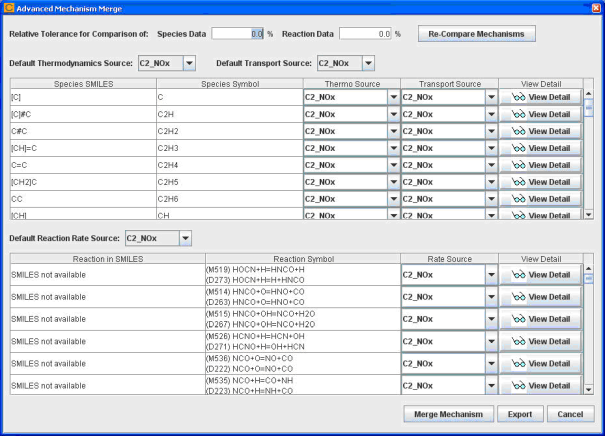
The Show Detailed Comparison option can be turned off if you do not anticipate fine-tuning these merge-precedence rules on a conflict-by-conflict basis. In this case, the merge will be completed using the global rules selected and the merged mechanism will be generated automatically. However, to look at the differences between full and donor mechanisms in detail and to resolve these conflicts explicitly, turn on the Show Detailed Comparison option. In this case a detailed comparison dialog will be displayed as shown in Figure 2.2: Detailed comparison dialog in Mechanism Merge . This dialog allows you to specify which mechanism's values should be used for each conflict if an override of the global rule is desired.
By default, if there is any difference at all in the data values from the two mechanisms, they will be treated as a conflict. For example, if the pre-exponential factor of the same reaction from the full mechanism is 1.00 while the value from the donor mechanism is 1.0001, this reaction will show up in the reaction table as a conflict. However, you might consider such a difference to be insignificant. In that case, you can set up a non-zero relative tolerance for the comparison of mechanism data. For example, if you set the relative tolerance to 1%, then a difference that is less than 1% of the value from the full mechanism will be ignored and the example reaction mentioned would not be identified as a conflict and the global rule for precedence would automatically take effect for that data.
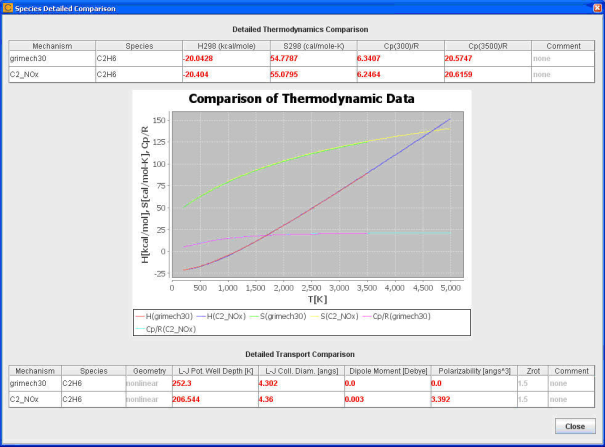
A View Detail button next to each conflict listed displays details regarding the differences found in the two mechanisms on a per-species or per-reaction basis. The details of thermodynamic and transport differences display on a per-species basis and the details, as shown in Figure 2-3, include:
Heat of formation at 298K.
Entropy of formation at 298K.
Heat capacity at the minimum and maximum of overlapped thermodynamic polynomials.
A plot of heat of formation, entropy of formation, and heat capacity calculated using the polynomials covering the corresponding temperature ranges.
Transport data.
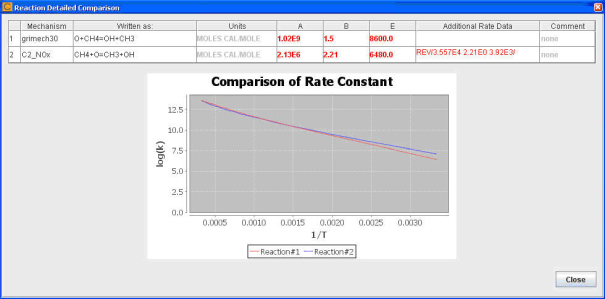
The details of a reaction conflict will include the reaction equation, as well as the rate parameters from the two mechanisms, as shown in Figure 2.4: Details of reaction rate differences for a reaction .
Once the merge operation is completed, you can view the results including:
The merged mechanism that has been created in the working directory.
The summary of the mechanism-merge operation, including the treatment of every reaction conflict between the mechanisms.
The summary of species mapping from donor mechanism to full mechanism, if SMILES assignments are available for every species.
The summary of missing SMILES assignments in the mechanisms, if applicable.
All reaction rates are included in the merged mechanism, but the reactions with conflicts in the mechanism are provided as comments that indicate how the conflict in the mechanism file was resolved; the version of the reaction that is no longer in use is commented out.