This is an optional file which can be used in conjunction with models
which have rotational or translational computational symmetry (or periodic boundary
conditions). It is invoked in the GEOMETRY section of the EnSight casefile, using the
"match: filename" line. EnSight 6 Case file allows the use of the periodic matchfile
which uses the matchfile in the border process to eliminate the matched symmetry faces
at the shared boundary between the faces. EnSight Case Gold allows the optional
[add_ghosts] parameter after the filename which will produce ghost
cells across the match file boundary which will provide continuity for variable
calculations across the boundary, and for computational symmetry/mirroring, etc. Only
use the add_ghosts option if you can afford the penalty of the additional
ghost cells and you need the computational continuity that they provide.
When a model piece is created with periodic boundary conditions, there is
usually a built-in correspondence between two faces of the model piece. If you transform
a copy of the model piece properly, face 1 of the copy will be at the same location as
face 2 of the original piece. It is desirable to know the corresponding nodes between
face 1 and face 2 so border elements will not be produced at the matching faces. This
correspondence of nodes can be provided in a periodic match file as indicated below.
(Please note that if a periodic match file is not provided, by default EnSight will
attempt to determine this correspondence using a float hashing scheme. This scheme has
been shown to work quite well, but may not catch all duplicates. The user has some
control over the "capture" accuracy of the hashing through the use of the command:
test: float_hash_digits. If this command is issued from the command
dialog, the user can change the number of digits, in a normalized scheme, to consider in
the float hashing. The lower the number of digits, the larger the "capture" distance,
and therefore the higher the number of digits, the smaller the capture distance. The
default is 4, with practical limits between 2 and 7 or 8 in most cases.)
The transformation type and delta value are contained in the file. The periodic match file is an ASCII free format file. For unstructured data, it can be thought of as a series of node pairs, where a node pair is the node number of face 1 and its corresponding node number on face 2. For structured blocks, all that is needed is an indication of whether the i, j, or k planes contain the periodic face. The min plane of this "direction" will be treated as face 1, and the max plane will be treated as face 2.
The file format is as follows:
rotate_x/y/z / translate |
The first line is either rotate_x, rotate_y, rotate_z or translate |
theta / dx dy dz |
The second line contains rotation angle in degrees or the three translational delta values. |
np n11 n21 n12 n22 . . . . . . n1np n2np |
If any unstructured pairs, the third line contains the number of these pairs (np). And the node ids of each pair follow. (The first subscript indicates face, the second is pair.) |
blocks bmin bmax i/j/k |
Last in the file comes as many of these "blocks" lines as needed. bmin and bmax are a range of block numbers. For a single block, bmin and bmax would be the same. Only one of i, j, or k can be specified for a given block. |
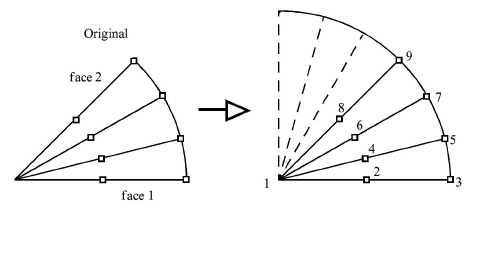
Simple unstructured rotational example:
The periodic match file for a rotation of this model about point 1 would be:
rotate_z
45.0
3
1 1
2 8
3 9
Thus, face 1 of this model is made up of nodes 1, 2, and 3 and face 2 of this model is made up of nodes 1, 8, and 9. So there are 3 node pairs to define, with node 1 corresponding to node 1 after a copy is rotated, node 2 corresponding to node 8, and node 3 corresponding to node 9.
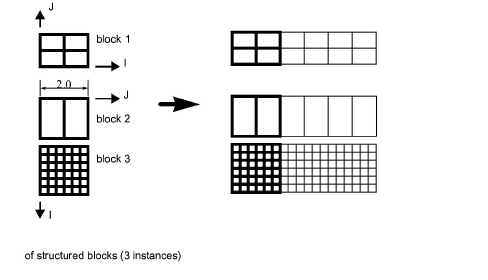
Simple structured translational model:
translate2.0 0.0 0.0blocks 1 1 i
blocks 2 3 j
Since block 1 is oriented differently than blocks 2 and 3 in terms of ijk space, two "blocks" lines were needed in the match file.
Special Notes / Limitations
This match file format requires that the unstructured node ids of the model be unique. This is only an issue with EnSight Gold unstructured format, since it is possible with that format to have non-unique node ids.
The model instance (which will be duplicated periodically) must have more than one element of thickness in the direction of the duplication. If it has only one element of thickness, intermediate instances will have all faces removed. If you have this unusual circumstance, you will need to turn off the shared border removal process, as explained in note 3.
The shared border removal process can be turned off, thereby saving some memory and processing time, by issuing the "test: rem_shared_bord_off" command in the command dialog. The effect of turning it off will be that border elements will be left between each periodic instance on future periodic updates.
The matching and hashing processes actually occur together. Thus, matching information does not have to be specified for all portions of a model. If no matching information is specified for a given node, the hashing process is used. By the same token, if matching information is provided, it is used explicitly as specified - even if it is has been specified incorrectly.